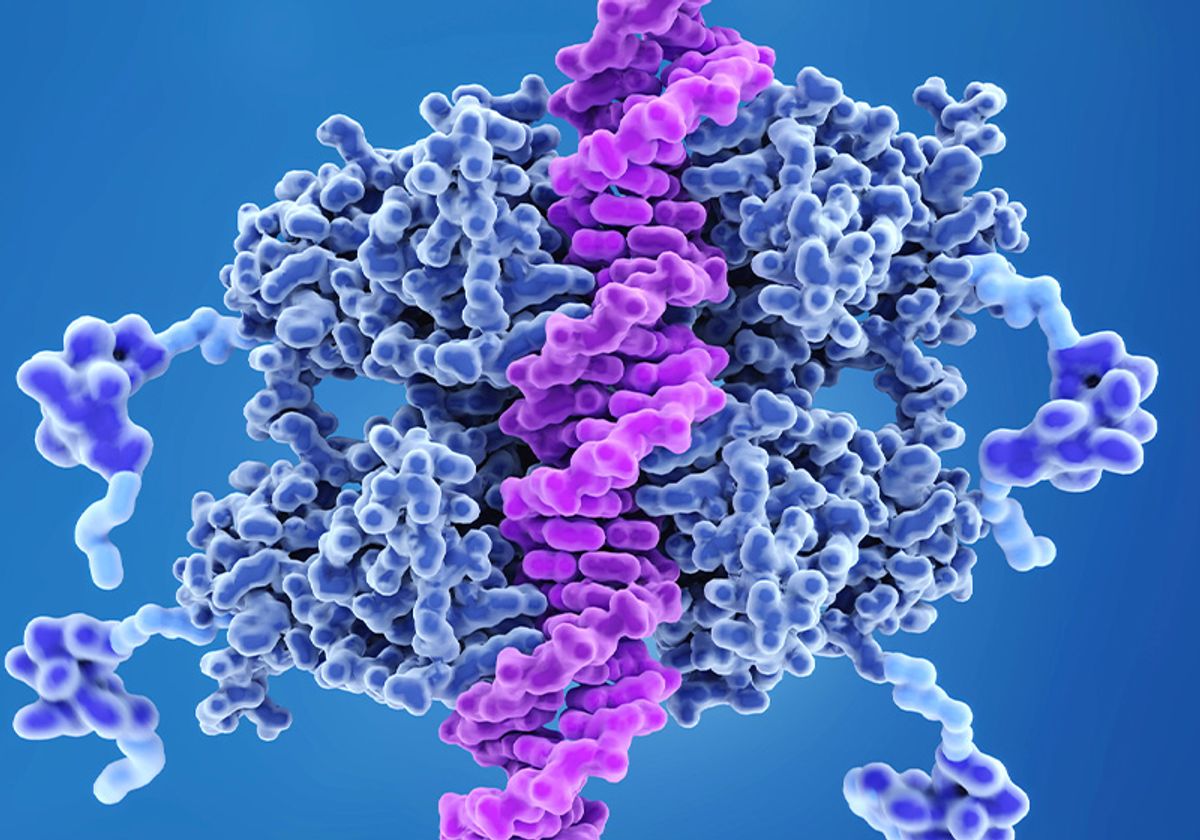
The tumor suppressor protein p53, known as the “guardian of the genome,” protects the body’s DNA from daily stress or long-term damage by triggering cells to either repair or self-destruct. But mutations in the p53 gene that codes for this protein prevent it from working, allowing errors to accumulate in the genetic code, leading to diseases such as cancer.
For the first time, a Penn State-led team has discovered the complete structure of the p53 protein using patient samples. They also investigated how mutation-induced changes in the structure of p53 affect different cancers. They published their findings in Chemical Biochemistry.
“We defined the full-length structure of p53, opening the door to understanding 3D arrangements that could be used for new therapies,” said Maria Solares, a PhD student in the Molecular, Cellular and Integrative Biosciences at Penn State’s Huck Institute for Life Sciences. She is also the lead author of the paper. “Scientists have previously identified p53 as an important focus of tumor origin and have studied its function in cells extensively. However, without understanding the full structure of p53, our knowledge about how it is processed in diseased cells is incomplete.”
The researchers used cryo-electron microscopy (cryo-EM) to image individual p53 proteins isolated from brain tumor cells. What’s unique about their work, Solares said, is that the team used a semiconductor material to cleanly capture the isolated p53 protein before imaging it. These silicon-based microchips hold proteins in such a way that researchers can resolve previously unseen molecular features.
“Seeing the full structure of p53 is like seeing only limited parts or limbs for a long time, then finally saw the whole human body.” “It’s hard to understand how they work without knowing their entire physical makeup.”
The p53 protein is made up of individual units called monomers, which join together to form larger entities, or dimers and tetramers.
“We found that within cancer cells there is a mixture of monomers, dimers and tetramers, each of which plays a different role depending on what’s going on in the nucleus,” Solares said. “The results of the dimer structure revealed for the first time the ‘closed’ structure of the molecule. This form of p53 is like the starting line before a race, ready to run to the DNA when other cell signaling goes wrong.”
Other forms of p53, such as monomers, are considered “open” because they need to bind to another unit in order to move to the starting line and perform tasks. Proper communication is important for controlling how much and when p53 deploys to the nucleus, where DNA resides, and disruptions in this process have been linked to the origins of diseases, including cancer, Kelly believes.
Mutations in the p53 gene cause the p53 protein to not communicate well, Kelly said. Once they revealed the 3D structure of p53, the researchers investigated how structural changes in p53 could cause the molecule to function improperly within cells.
“It is well known that half of all cancers contain mutations in the p53 gene,” Solares said. “To further investigate how these mutations affect the structure of the p53 protein, we used molecular modeling software to simulate changes in the structure of the p53 monomer.”
The researchers looked at seven “hotspots,” where mutations in the protein’s structure are most often associated with cancer. These seven mutations in the p53 protein often led to less favorable patient outcomes in terms of disease progression and chemotherapy resistance, Kelly said.
They found that slight changes in the 3D structure of mutant p53 affect the protein’s surface charges, which can repel and attract charges from other molecular units. According to Kelly, this could hamper the normal interactions between proteins and DNA, leading to disruption of p53’s ability to assist in regulatory or repair processes necessary to maintain healthy cells.
“Under healthy conditions, p53 uses zinc ions to hold onto genetic material,” Solares said. “When the protein’s surface charge is properly balanced, zinc ions can be in the right position to help p53 hold onto DNA. But when mutations in the p53 gene cause changes on the surface of the protein, the zinc ions are not placed correctly, and p53 loses control of the DNA. This effect is further exacerbated in disease.”
In the future, the researchers say they hope to extend their work to pancreatic and ovarian cancers that are strongly associated with p53 mutations. They are also working on new treatments based on their new understanding of the full 3D structure of p53.
“One patient’s cancer is not the same as another patient’s cancer,” Solares said. “We need to keep collecting experimental data from different patients and different cancers to test our models. We can’t fully understand cancer without the full picture.”