An unusual protein structure called a “corrugated beta sheet”, first predicted in 1953, has now been created in the laboratory and characterized in detail using X-ray crystallography.
The new findings, published in Chemical Science in July, may enable the rational design of unique materials based on corrugated sheet-like structures.
Jevgenij Raskatov, associate professor of chemistry and biochemistry at the University of California, Santa Cruz and corresponding author of the paper, said: “Our study establishes the corrugated beta sheet configuration as a motif with universal characteristics, and opens the way for structure-based design of unique molecular structures with the potential for material development and biomedical applications.”
Proteins come in ever-changing shapes and sizes and play a wide variety of structural and functional roles in living cells. Certain common structural motifs, such as alpha helices, are found in many protein structures.
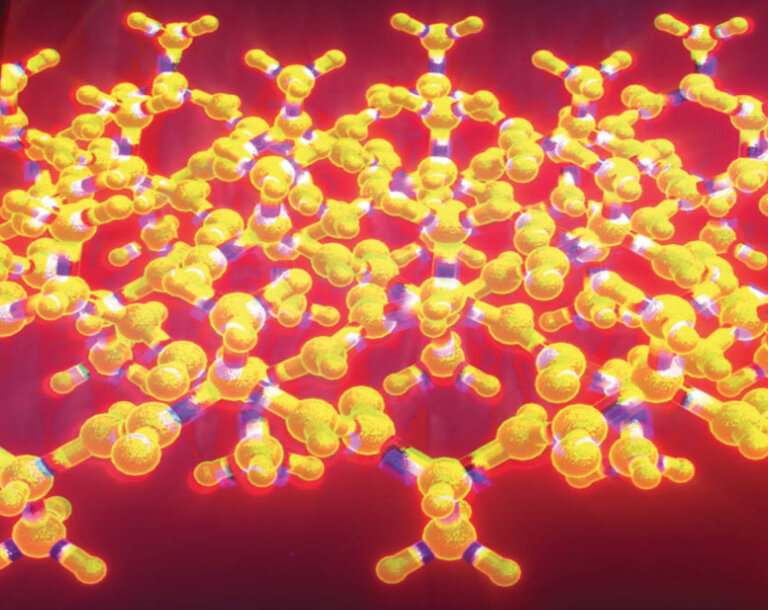
Corrugated sheet is a variation of β sheet, which is a famous structural motif found in thousands of proteins. Linus Pauling and Robert Corey described corrugated beta flakes in 1953, and two years ago they proposed the concept of corrugated beta flakes. Although the folded beta sheet is well-known and often simply called beta sheet, corrugated beta sheets have remained largely a theoretical structure for decades.
In a study previously published in 2021, Raskatov’s team reported that by mixing small peptides with an equal amount of mirror images, they obtained corrugated beta sheet structures. The researchers used the mirror-image form of triphenylalanine, a short peptide composed of three phenylalanine amino acids. The mirror-image polypeptides bind in pairs to form “dimers” with the predicted structure, but they do not form the extended, periodically corrugated beta-sheet topography assumed by Pauling and Corey.
“The dimers clustered together to form a chevron-shaped layer structure, which raised doubts about whether a periodically corrugated beta-sheet structure was feasible,” Raskatov said.
In the new study, the researchers replaced one of the triphenylalanines with other amino acids, resulting in a slightly different tripeptide and its mirror image. Using these new tripeptides, they were able to create three different polymeric peptide systems that form extended layers of antiparallel corrugated beta sheets in which mirror-image peptide chains are arranged in an alternating fashion. The X-ray crystallography results showed that the crystal structure was basically consistent with Pauling and Corey’s predictions.
One of the paper’s first authors is Raskatov’s lab staff Amaruka Hazari and UCLA’s Michael Sawaya. Other co-authors include Timothy Johnstone of UC Santa Cruz and Niko Vlahakis, David Boyer, Jose Rodriguez and David Eisenberg of UCLA. This research was supported by the National Institutes of Health.